B-hLUCA2 mice
| Strain Name |
C57BL/6N-Luca2tm1(LUCA2)Bcgen/Bcgen
|
Common Name | B-hLUCA2 mice |
| Background | C57BL/6N | Catalog number |
112499 |
|
Aliases |
Human: HYAL2 | ||
Description
- HYAL2, a glycosylphosphatidylinositol (GPI)-anchored membrane receptor, binds and cleaves HMW-HA into low molecular-weight HA (LMW-HA; ~ 20 kDa) fragments at the cell membrane. This mechanism has been proposed to require the presence and activity of the major HA receptor CD44, which is essential for intracellular signaling. Inside lysosomes, HA is further processed into oligomeric HA fragments by HYAL1, together with exoglycosidases.
- Hyaluronidase (HYAL)-2 has been identified as one of the principal enzymes involved in HA catabolism in vertebrates. HYAL2 is broadly expressed in tissues but has catabolic function within only a narrow acidic pH range (optimal pH, 4), and compared to other HYALs, has only weak intrinsic HA-degrading activity.
- HYAL2 was originally identified as a lysosomal enzyme but was also subsequently identified as anchored to the cell membrane via a glycosylphosphatidylinositol link.15 Aberrant expression of HYAL2 is implicated in diverse pathology, including cardiac and skeletal abnormalities, hematopoietic and platelet dysfunction, cancer, and fibrosis.
Mechanism of action
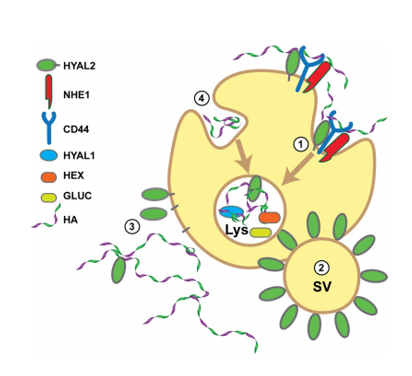
Proposed model of HA degradation. HA degradation occurs through more than one pathway, each indicated by different numbers, 1–4, in this model.
In pathway 1, cell surface HYAL2 initiates the hydrolysis of HA in partnership with a receptor, possibly CD44. NHE1 acidifies the local region to allow HYAL2 cleavage. The resulting HA fragments are endocytosed and transported to the lysosome (Lys) where they are degraded through the action of the endoglycosidase HYAL1 and the exoglycosidases, β-hexosaminidase (HEX) and β-glucuronidase (GLUC).
In pathway 2, secretory vesicles (SV) within specialized epithelial cells are surrounded by HYAL2. Upon secretion, these vesicles will degrade extracellular HA.
In pathway 3, cell surface HYAL2 is shed from the surface or released through by a lipase or protease to form a soluble form of HYAL2. This enzyme can enter the blood stream or diffuse from the surface to degrade HA distal from the site of HYAL2 expression.
In pathway 4, HA is internalized by a non-HYAL2-dependent route and transported to the lysosome for degradation.
Gene targeting strategy for B-hLUCA2 mice.
The exons 2~4 of mouse Luca2 gene that encode the full-length protein were replaced by human LUCA2 exons 2~4 in B-hLUCA2 mice.
Model validation
Protein expression analysis

mRNA expression analysis
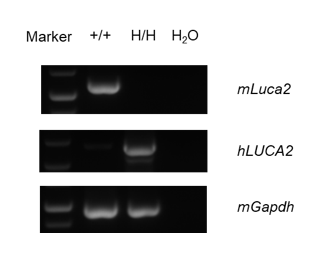
Strain specific analysis of LUCA2 mRNA expression in wild-type C57BL/6 mice and B-hLUCA2 mice by RT-PCR. Kidney RNA were isolated from wildtype C57BL/6 mice (+/+) and homozygous B-hLUCA2 mice (H/H), then cDNA libraries were synthesized by reverse transcription, followed by PCR with mouse or human LUCA2 primers. Mouse Luca2 mRNA was detectable only in wild-type C57BL/6 mice. Human LUCA2 mRNA was detectable only in homozygous B-hLUCA2 mice but not in wild-type mice.
Summary
Protein expression analysis:
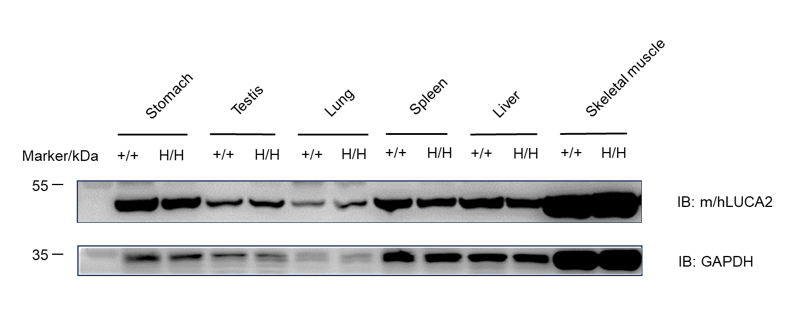
LUCA2 protein was detected in stomach, testis, lung, spleen, liver and skeletal muscle of wild-type C57BL/6 mice and homozygous B-hLUCA2 mice.










